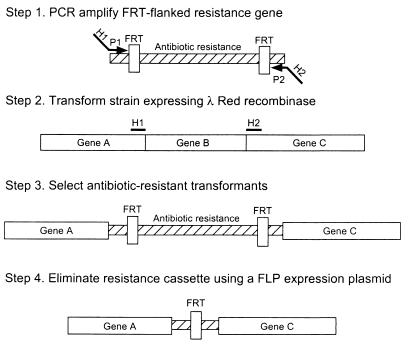
One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products.
We have developed a easy and extremely environment friendly technique to disrupt chromosomal genes in Escherichia coli in which PCR primers present the homology to the focused gene(s).
In this process, recombination requires the phage lambda Red recombinase, which is synthesized underneath the management of an inducible promoter on an simply curable, low copy quantity plasmid.
To display the utility of this method, we generated PCR merchandise by using primers with 36- to 50-nt extensions which can be homologous to areas adjoining to the gene to be inactivated and template plasmids carrying antibiotic resistance genes which can be flanked by FRT (FLP recognition goal) websites.
By using the respective PCR merchandise, we made 13 completely different disruptions of chromosomal genes. Mutants of the arcB, cyaA, lacZYA, ompR-envZ, phnR, pstB, pstCA, pstS, pstSCAB-phoU, recA, and torSTRCAD genes or operons have been remoted as antibiotic-resistant colonies after the introduction into micro organism carrying a Red expression plasmid of artificial (PCR-generated) DNA.
The resistance genes have been then eradicated by using a helper plasmid encoding the FLP recombinase which can also be simply curable. This process ought to be extensively helpful, particularly in genome evaluation of E. coli and different micro organism as a result of the process will be finished in wild-type cells.
The MIQE tips: minimal info for publication of quantitative real-time PCR experiments.
BACKGROUNDCurrently, an absence of consensus exists on how greatest to carry out and interpret quantitative real-time PCR (qPCR) experiments. The drawback is exacerbated by an absence of enough experimental element in many publications, which impedes a reader’s means to judge critically the standard of the outcomes introduced or to repeat the experiments.
BACKGROUNDThe Minimum Information for Publication of Quantitative Real-Time PCR Experiments (MIQE) tips goal the reliability of outcomes to assist make sure the integrity of the scientific literature, promote consistency between laboratories, and improve experimental transparency.
MIQE is a set of tips that describe the minimal info crucial for evaluating qPCR experiments. Included is a guidelines to accompany the preliminary submission of a manuscript to the writer.
By offering all related experimental situations and assay traits, reviewers can assess the validity of the protocols used. Full disclosure of all reagents, sequences, and evaluation strategies is critical to allow different investigators to breed outcomes.
MIQE particulars ought to be printed both in abbreviated type or as a web-based complement.
CONCLUSIONSFollowing these tips will encourage higher experimental observe, permitting extra dependable and unequivocal interpretation of qPCR outcomes.
