Cusabio Arabidopsis thaliana Recombinant
Abstract
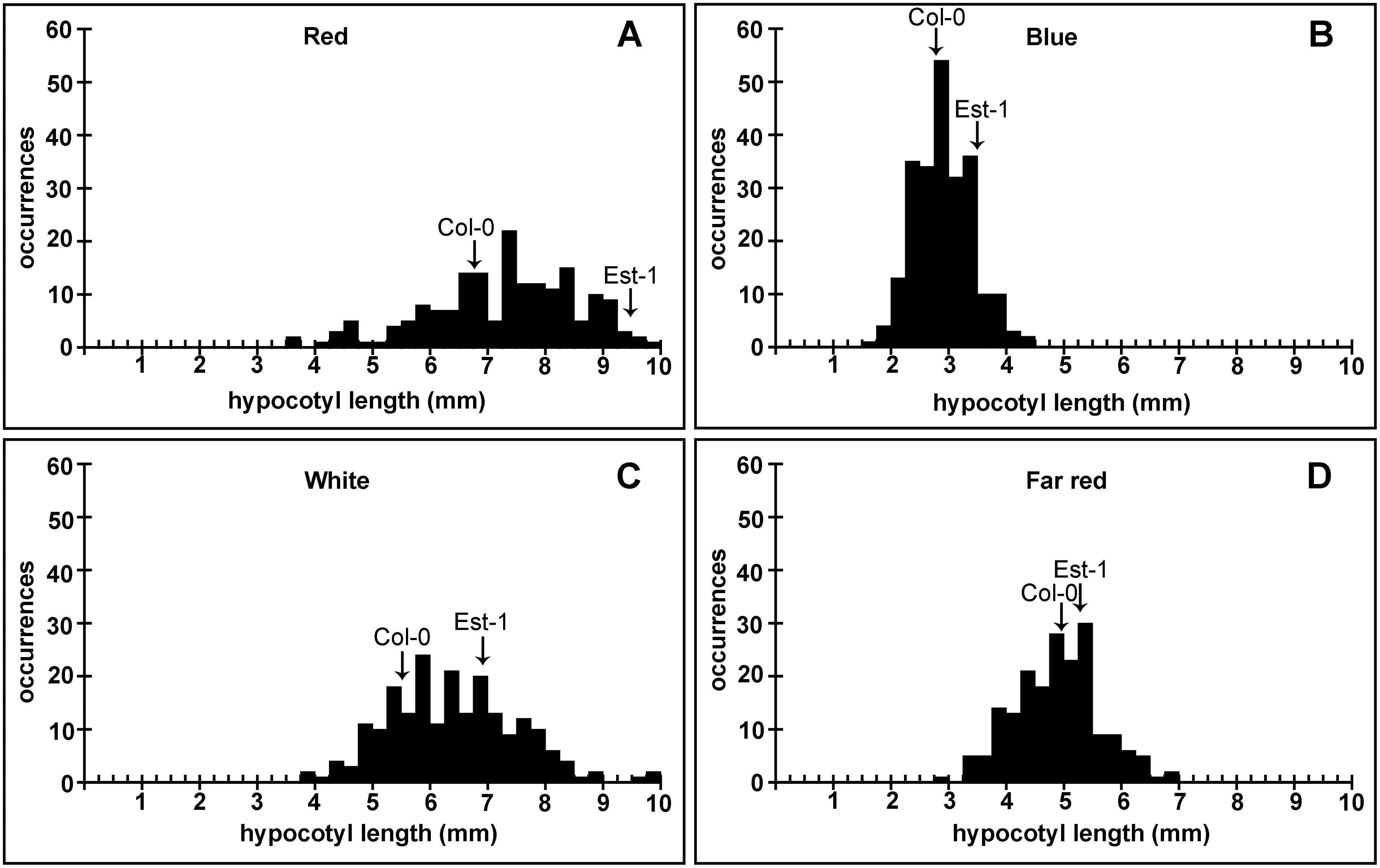
Recombinant populations were the basis of Mendel’s early genetic experiments and continue to be key to the study of genes, heredity, and genetic variation today. Genotyping several hundred thousand loci in a single assay by hybridizing genomic DNA to oligonucleotide arrays provides a powerful technique for improving precision linkage mapping. The genotypes of two Arabidopsis thaliana Recombinant accessions were compared using a 400,000-feature exon-specific oligonucleotide array.
Approximately 16,000 single feature polymorphisms (SFPs) were detected in approximately 8,000 of the approximately 26,000 genes represented on the array. Allelic variation at these loci was measured in a population of recombinant inbred lines, which defined the location of 815 recombination breakpoints. The genetic linkage map had a total length of 422.5 cm, with 676 informative SFP markers representing ~0.6 cM intervals. One hundred and fifteen single gene intervals were identified.
The recombination rate, SFP distribution, and segregation in this population are not uniform. Many genomic regions show a cluster of recombination events that include significant hot spots. The precise haplotype structure of the recombinant population was defined with unprecedented precision and resolution. The resulting linkage map allows further refinement of the hundreds of quantitative trait loci identified in this well-studied population. Highly variable recombination rates along each chromosome and extensive segregation distortion was observed in the population.
Purity: greater than 85% as determined by SDS-PAGE.
Destination Names: NLP7
Uniprot No.: Q84TH9
Research Area: Epigenetic and Nuclear Signaling
Alternative Names: NIN-like protein 7
Species: Arabidopsis thaliana (Mouse-ear cress)
Source: Baculovirus
Expression Region: 1-959aa
Mole Weight: 107.4 kDa
Protein Length: Total length
Tag Information: N-terminal 10xHis-tagged
Form: Liquid or Lyophilized Powder
Note: We will preferably ship the format we have in stock, however, if you have any special requirements for the format, please remark your requirement when placing the order, we will prepare according to your demand.
Buffer
If the dosage form is liquid, the default storage buffer is a Tris/PBS based buffer, 5%-50% glycerol. If the administration form is a lyophilized powder, the buffer prior to lyophilization is a Tris/PBS based buffer, 6% trehalose, pH 8.0.
Reconstitution
We recommend that this vial be briefly centrifuged before opening to bring the contents to the bottom. Reconstitute protein in sterile deionized water at a concentration of 0.1-1.0 mg/mL. We recommend adding 5-50% glycerol (final concentration) and an aliquot for long-term storage at -20°C/-80°C. Our final default glycerol concentration is 50%. Customers could use it for reference.
Storage Conditions
Store at -20°C/-80°C upon receipt, need to be aliquoted for multiple uses. Avoid repeated cycles of freezing and thawing.
Shelf life
Shelf life is related to many factors, storage condition, buffer ingredients, storage temperature and the stability of the protein itself. Generally, the shelf life of the liquid form is 6 months at -20°C/-80°C. The shelf life of the lyophilized form is 12 months at -20°C/-80°C.
Delivery time: 3-7 business days
Notes: Repeated freezing and thawing is not recommended. Store working aliquots at 4°C for up to one week.
Synopsis
One goal of many genetic studies is to discover the underlying genetic condition (the genotype) of a specific physical manifestation in an organism (the phenotype), such as diabetes in humans or leaf rust in cultivated wheat. One limitation to making such discoveries is the ability to resolve the genotype. Gene arrays carry representations of the genome, called features, in high density on a thumbnail-sized surface.
In this study, microarrays designed to measure gene expression were used to detect polymorphisms in the DNA sequence. DNA from two different Arabidopsis strains was hybridized to arrays representing almost the entire coding region of the genome. Differences in hybridization intensity indicated differences in DNA sequence. Sequence differences, termed single trait polymorphisms, were then analyzed in a population of 100 plants derived from inbreeding of the progeny of the two-parent strains.
The precise location of the genetic recombination breakpoints was defined for each line. As a result, Singer et al. were able to generate one of the first very high-resolution genotyping datasets in a multicellular organism that allowed the construction of a high-resolution genetic map of Arabidopsis. This map will greatly facilitate attempts to make definitive associations between genotypes and phenotypes.
Vegetal material.
Columbia/Landsberg RIL (Col/Ler) seeds (eight generations inbreeding) [13] and parental lines were kindly provided by the Arabidopsis Biological Resource Center (ABRC) at The Ohio State University, Columbus, Ohio, United States. The accessions used in this study correspond to the first set of 100 RIL (CS1899), the parental lines were Columbia (CS933; Col-4, named Col) and Landsberg erecta (CS20, named Ler). Eight plants for each RIL were grown in a pot under long-day conditions (16 h light, 8 h dark) and pooled for analysis.
DNA isolation, labelling and hybridization of microarrays.
Total genomic DNA was isolated from leaf tissue with the DNeasy Plant Mini Kit (Qiagen, Valencia, California, United States) according to the manufacturer’s instructions. Four and six biological replicates of Col (CS933) and Ler (CS20) were made, respectively. For RILs 1 (CS1900) to 57 (CS1957), a single replicate was available, except for lane 6 (CS1906) which was tripled.
RILs 58 (CS1958) to 100 (CS4686) were doubled, except that line 73 (CS1973) was tripled and lines 83 (CS1983), 84 (CS1984) and 89 (CS1989) were quadrupled. Probe intensity values for replicate RIL microarrays were averaged for genotyping. DNA was labelled by random priming with biotin14-dCTP (Bioprime DNA Labeling System, Invitrogen, Carlsbad, California, USA). Hybridization, washing, staining and scanning were carried out using the standard Affymetrix Eukaryotic protocol. GeneChip Suite was used for image acquisition.